Scientists use artificial intelligence to find drugs to fight drug-resistant infections
Machine learning algorithm identifies a compound that can kill people Acinetobacter baumanniia bacterium that lurks in many hospital settings.
Researchers at MIT and McMaster University have used an artificial intelligence algorithm to discover a new antibiotic that kills a bacterium that causes many drug-resistant infections.
If developed for use in patients, the drug could help fight Acinetobacter baumannii, a bacterium often found in hospitals that can cause pneumonia, meningitis and other serious infections. This microorganism is also a leading cause of infection among wounded soldiers in Iraq and Afghanistan.
“Acinetobacter can survive on doorknobs and hospital equipment for long periods of time and can acquire antibiotic resistance genes from its environment. It is now common to find isolates of Acinetobacter baumannii that are resistant to almost all antibiotics, ” said Jonathan Stokes, a former MIT postdoc and current assistant professor of biochemistry and biomedical sciences at McMaster University.
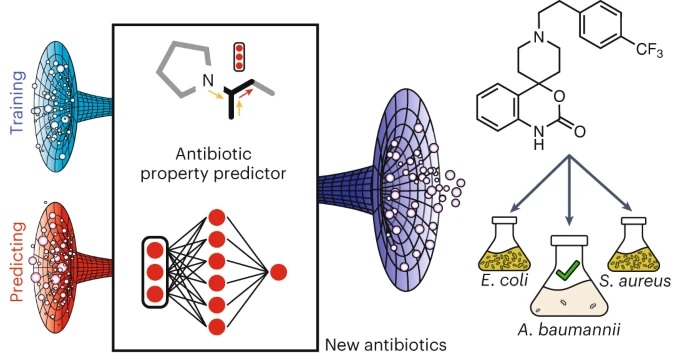
The researchers identified the new drug from a library of nearly 7,000 potential drug compounds using a machine learning model they trained to evaluate whether compounds would inhibit the growth of Acinetobacter baumannii.
“This discovery further supports the premise that artificial intelligence can significantly accelerate and expand our search for new antibiotics,” said James Temeer Professor of Medical Engineering and Science in the Institute of Medical Engineering and Science (IMES) and the Department of Bioengineering James Collins said. MIT. “I’m delighted that this work shows that we can use artificial intelligence to help combat problematic pathogens like Acinetobacter baumannii.”
Collins and Stokes are lead authors of the new study, published today in Nature Chemical Biology. The lead authors of the paper are McMaster graduate students Gary Liu and Denise Catacutan, and recent McMaster graduate Khushi Rathod.
drug discovery
Over the past few decades, many pathogenic bacteria have become increasingly resistant to existing antibiotics, while new antibiotics are rarely developed.
A few years ago, Collins, Stokes and MIT professor Regina Barzilay (also an author of the new study) set out to address this growing problem by using machine learning, which is a type of artificial intelligence that can learn to recognize patterns in the environment. Large amounts of data. Collins and Barzilay, co-directors of MIT’s Abdul Latif Jameel Health Machine Learning Clinic, hope this approach can be used to identify new antibiotics whose chemical structures are different from any existing drugs.
In an initial demonstration, the researchers trained a machine learning algorithm to identify chemical structures that inhibit the growth of E. coli.In a screen of more than 100 million compounds, the algorithm produced a molecule that the researchers dubbed halicin, after the fictional artificial intelligence system “2001: A Space Odyssey”. They demonstrated that the molecule could kill not only E. coli but also several other types of bacteria that were resistant to this treatment.
“Following that paper, when we showed that these machine learning methods worked well at the complex task of antibiotic discovery, we turned our attention to what I consider the number one public enemy of multidrug-resistant bacterial infections, Acinetobacter.”
To obtain training data for their computational model, the researchers first exposed Acinetobacter baumannii growing in lab dishes to about 7,500 different compounds to see which ones inhibited the microbe’s growth. They then input the structure of each molecule into the model. They also told the model whether each structure could inhibit bacterial growth. This enables the algorithm to learn chemical features associated with growth inhibition.

